
web-code version: 2019-11-04
database version: 27.01
Tweets by South Green
 Contact us.
Contact us.
Enter a gene name
You can restrict the search to one species (ancestral or modern).
Selected examples: Seita.3G233000.1, Sobic.009G148000.1, WRKY
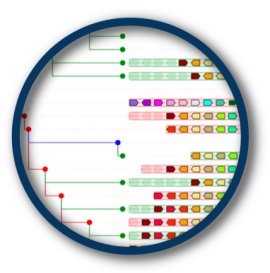
Genomicus is a genome browser that enables users to navigate in genomes in several dimensions: linearly along chromosome axes, transversaly across different species, and chronologicaly along evolutionary time.
Once a query gene has been entered, it is displayed in its genomic context in parallel to the genomic context of all its orthologous and paralogous copies in all the other sequenced metazoan genomes. Moreover, Genomicus stores and displays the predicted ancestral genome structure in all the ancestral species within the phylogenetic range of interest.
| Number of extant species | 27 |
|---|---|
| Number of extant genes | 1040086 |
| Number of ancestral species | 17 |
| Number of ancestral genes | 810245 |
| Number of ancestral synteny blocks | 0 |